clear all
The following lines of code add a path to all the Bayesian statistical functions.
% Add MCMC codes to path current_directory = cd; addpath(strcat(current_directory,'/mcmcstat'));
The following analysis computes two Landau energy parameters used in estimating changes in energy calculated from prior density functional theory (DFT) calculations. The DFT calculations were computed by incrementing atoms in lead titanate in a prescribed manner to produce changes in polarization about a fixed cubic strain state. Thus, only the polarization energy is required for model calibration. The effect of electrostriction is treated differently using a separate model for stress. More details about this analysis can be found in:
Oates, W., "A Quantum-informed Continuum Model for Ferroelectric Materials," Smart Mater. Struct. v. 23(10), p. 104009, 2014.
The form of the energy modeled considered for model calibration is
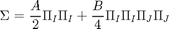
where 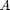 and
and 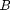 are the Landau parameters that will be calibrated through the Bayesian analysis. The vector order parameter is denoted by
are the Landau parameters that will be calibrated through the Bayesian analysis. The vector order parameter is denoted by 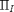 with
with 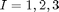 and is proportional to polarization.
and is proportional to polarization.
Define a function that calculates the sum of squares of error between the DFT energy and the approximation based on the Landau continuum energy function.
ssfun = @coupled_func_Bayesian; model.ssfun = ssfun;
Load the DFT calculations to fit the continuum scale Landau energy. A scaling parameter is introduced (1e3) that converts the energy from GPa units to MPa units. This scaling is applied on the initial parameter estimates and their bounds if the user wants to use a different set of units in fitting Landau energy parameters.
load DFT_cubic model.N = length(Pnum); %length of the different polarization and energy states scaling = 1e3; %all DFT energy density is in units of MPa using the scaling 1e3 %cubic volume V0 = 4.1476034E+02; %volume of DFT lattice in Bohr^3 V1 = V0*(5.29177249e-11)^3; %conversion of volume to m^3 deltaE = (E_tot - E_tot(1))/V1*1.60217656535e-19/1e9; %Giga-Joules/m^3 conversion from eV/m^3 %input energy paramters from DFT data.ydata_E_tot = deltaE*scaling; data.xdata = Pnum; %polarization computed from DFT using the Berry phase approach %define initial guess for Landau parameters A = -0.805*scaling; B = 3.32*scaling; tmin = [A B]; %Landau energy model parameter range and initial guess params = { {'A', tmin(1), -5e6*scaling, 5e6*scaling} {'B', tmin(2), -5e3*scaling,5e8*scaling} };
Check model initial output prior to running a parameter estimation using MCMC
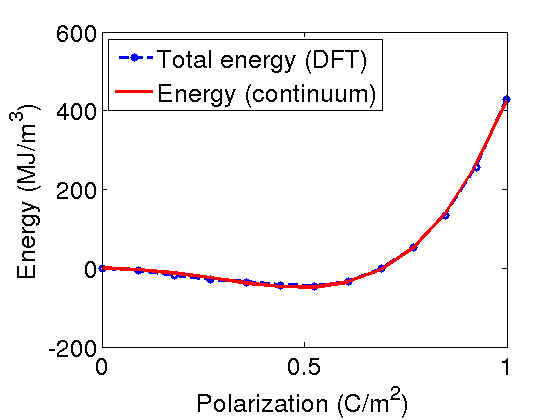
[Sigma] = coupled_model_Bayesian(tmin,data.xdata); ss = coupled_func_Bayesian(tmin,data); SS = ss/length(data.xdata); %here is the cost calculation figure(1) axes1 = axes('FontSize',20); plot(Pnum,data.ydata_E_tot,'bo--','Linewidth',3) hold on plot(Pnum,Sigma,'r-','Linewidth',3) hold off ylabel('Energy (MJ/m^3)','FontSize',20) xlabel('Polarization (C/m^2)','FontSize',20) legend('Total energy (DFT)','Energy (continuum)','Location','NorthWest') legend1 = legend(axes1,'show'); set(legend1,'EdgeColor',[1 1 1],'YColor',[1 1 1],'XColor',[1 1 1]); variance = sum((data.ydata_E_tot-Sigma').^2)/(length(deltaE-1)); %calculate the variance
Run the Bayesian statistical analysis. An initial run with a smaller number of iterations is first done and used as initial results in a longer set of iterations.
model.S20 = variance;%estimate of variance based on prior simulation model.N0 = 1; model.N = length(Pnum); options.updatesigma=1; options.method = 'dram'; %this applies the DRAM algorithm options.nsimu = 500; %run an initial small set of iterations prior to larger run [results, chain, s2chain] = mcmcrun(model,data,params,options); options.nsimu = 10000; %run longer simulations using results from smaller run as %initial conditions [results, chain, s2chain] = mcmcrun(model,data,params,options,results);
Setting nbatch to 1 Sampling these parameters: name start [min,max] N(mu,s^2) A: -805 [-5e+09,5e+09] N(0,Inf) B: 3320 [-5e+06,5e+11] N(0,Inf) Setting nbatch to 1 Using values from the previous run Sampling these parameters: name start [min,max] N(mu,s^2) A: -787.005 [-5e+09,5e+09] N(0,Inf) B: 3272.77 [-5e+06,5e+11] N(0,Inf)
Plot results.
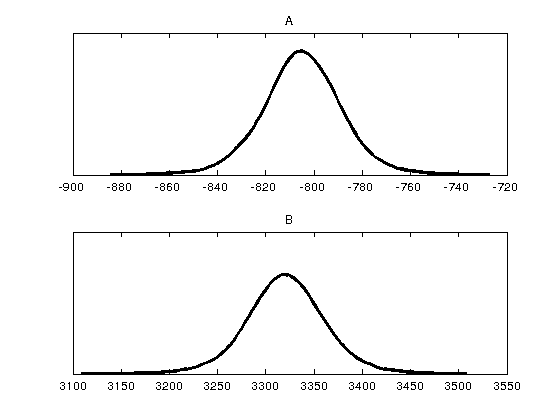
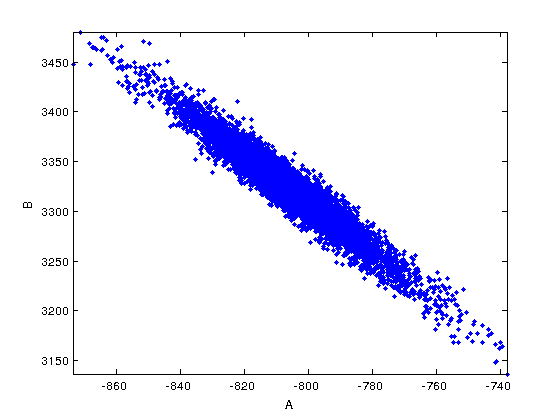
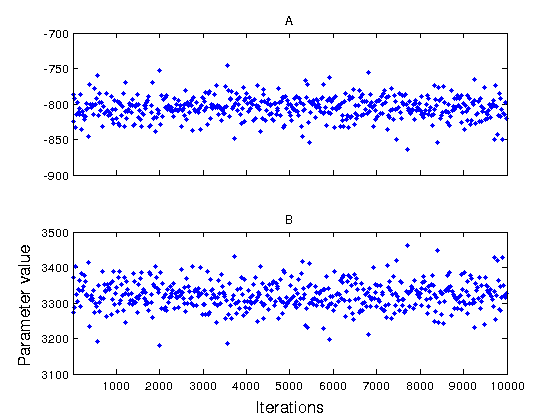
figure(2) mcmcplot(chain,[],results,'denspanel',2); figure(3) mcmcplot(chain,[],results,'pairs'); figure(4); clf mcmcplot(chain,[],results.names,'chainpanel') xlabel('Iterations','Fontsize',14) ylabel('Parameter value','Fontsize',14) chainstats(chain,results) %summary of parameter means, standard deviations, etc.
MCMC statistics, nsimu = 10000
mean std MC_err tau geweke
---------------------------------------------------------------------
A -804.99 16.144 0.5056 6.9381 0.99921
B 3320.7 40.049 1.1616 6.974 0.99928
---------------------------------------------------------------------
Define a model function that calls the chains for error propagation that give calculation of prediction and credible intervals
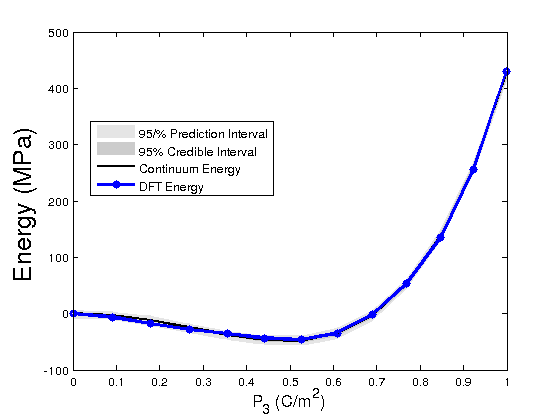
modelfun1 = @(d,th)coupled_model_Bayesian(th,d); % NOTE: the order in which d and th appear is important %The following lines of code propagate the posterior densities of %parameters to determine prediction and credible intervals of the Landau %energy with respect to the DFT energy in light of the Bayesian statistics nsample = 500; out = mcmcpred(results,chain,s2chain,data.xdata,modelfun1,nsample); figure(5) modelout = mcmcpredplot(out); %,data hold on plot(data.xdata,data.ydata_E_tot,'bo-','linewidth',3) hold off xlabel('P_{3} (C/m^2)','Fontsize',14); ylabel('Energy (MPa)','FontSize',20) legend('95/% Prediction Interval','95% Credible Interval','Continuum Energy','DFT Energy','Location','Best')